TreeView X is an open source and multi-platform program to display phylogenetic trees.
It can read and display NEXUS and Newick format tree files (such as those output by PAUP*, ClustalX, TREE-PUZZLE, and other programs). It allows users to order the branches of the trees, and to export the trees in SVG format.
It uses the wxWidgets C++ library.
Features include:
- Tree editor:
- Slanted cladogram.
-
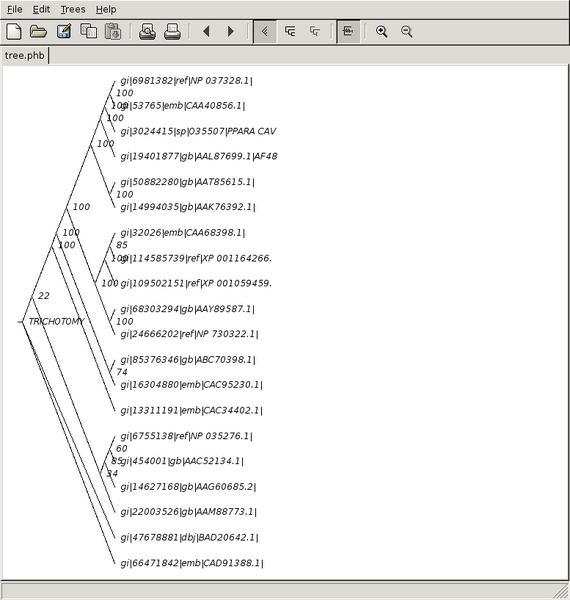
- Rectangular cladogram.
-
- Phylogram.
- Show internal node labels.
- Zoom.
- Reads many different tree file formats (including NEXUS, PHYLIP, Hennig86, NONA, MEGA, and ClustalW/X).
- Support for picture files (SVG and EMF).
- Export to SVG format.
- Print preview.
- Print multiple trees per page, and one tree over more than one page.
- Drag-and-drop.
Website: code.google.com/archive/p/treeviewx
Support:
Developer: Roderic D. M. Page
License: GNU General Public License v2.0
TreeView X is written in C++. Learn C++ with our recommended free books and free tutorials.
| Popular series | |
|---|---|
| The largest compilation of the best free and open source software in the universe. Each article is supplied with a legendary ratings chart helping you to make informed decisions. | |
| Hundreds of in-depth reviews offering our unbiased and expert opinion on software. We offer helpful and impartial information. | |
| The Big List of Active Linux Distros is a large compilation of actively developed Linux distributions. | |
| Replace proprietary software with open source alternatives: Google, Microsoft, Apple, Adobe, IBM, Autodesk, Oracle, Atlassian, Corel, Cisco, Intuit, SAS, Progress, Salesforce, and Citrix | |
| Awesome Free Linux Games Tools showcases a series of tools that making gaming on Linux a more pleasurable experience. This is a new series. | |
| Machine Learning explores practical applications of machine learning and deep learning from a Linux perspective. We've written reviews of more than 40 self-hosted apps. All are free and open source. | |
| New to Linux? Read our Linux for Starters series. We start right at the basics and teach you everything you need to know to get started with Linux. | |
| Alternatives to popular CLI tools showcases essential tools that are modern replacements for core Linux utilities. | |
| Essential Linux system tools focuses on small, indispensable utilities, useful for system administrators as well as regular users. | |
| Linux utilities to maximise your productivity. Small, indispensable tools, useful for anyone running a Linux machine. | |
| Surveys popular streaming services from a Linux perspective: Amazon Music Unlimited, Myuzi, Spotify, Deezer, Tidal. | |
| Saving Money with Linux looks at how you can reduce your energy bills running Linux. | |
| Home computers became commonplace in the 1980s. Emulate home computers including the Commodore 64, Amiga, Atari ST, ZX81, Amstrad CPC, and ZX Spectrum. | |
| Now and Then examines how promising open source software fared over the years. It can be a bumpy ride. | |
| Linux at Home looks at a range of home activities where Linux can play its part, making the most of our time at home, keeping active and engaged. | |
| Linux Candy reveals the lighter side of Linux. Have some fun and escape from the daily drudgery. | |
| Getting Started with Docker helps you master Docker, a set of platform as a service products that delivers software in packages called containers. | |
| Best Free Android Apps. We showcase free Android apps that are definitely worth downloading. There's a strict eligibility criteria for inclusion in this series. | |
| These best free books accelerate your learning of every programming language. Learn a new language today! | |
| These free tutorials offer the perfect tonic to our free programming books series. | |
| Linux Around The World showcases usergroups that are relevant to Linux enthusiasts. Great ways to meet up with fellow enthusiasts. | |
| Stars and Stripes is an occasional series looking at the impact of Linux in the USA. | |
